Single-stranded DNA for Gene Editing
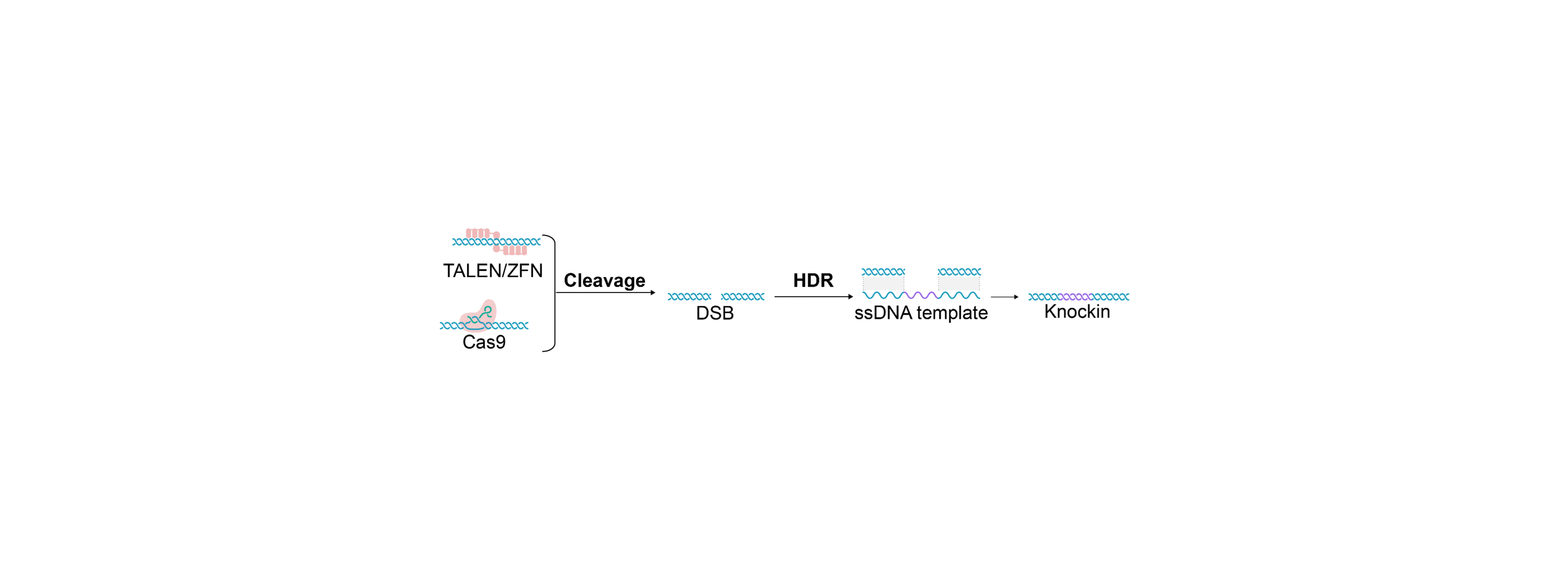
Several publications demonstrate that single-stranded DNA offers distinct advantages in gene editing, including lower cell toxicity, reduced off-target integrations, and higher knock-in efficiency. Single-stranded DNA provides significant benefits when used as a donor template in homology-directed repair mechanisms alongside gene editing tools like CRISPR/Cas. This is particularly advantageous for primary cells, such as T-cells, which are highly sensitive to double-stranded DNA.
Cell Therapy
Leverage the power of single-stranded DNA to advance targeted cancer therapies involving large gene insertions. Single-stranded DNA enables precise and efficient integration of T-cell receptors (TCRs) or chimeric antigen receptors (CARs) in cell therapies while minimizing cellular toxicity, ensuring higher viability and functionality.
Gene Therapy
Unlock the potential of single-stranded DNA in addressing genetic disorders like cystic fibrosis, sickle cell anemia, and Duchenne muscular dystrophy. Single-stranded DNA serves as a precise and reliable template for gene replacement and insertion therapies, offering transformative solutions for patients.
Agriculture and Food Technology
Revolutionize crop genomes with single-stranded DNA to enhance pathogen resistance, boost productivity, and increase resilience to contribute to sustainable agriculture and global food security.
Industrial Biotechnology
Optimize microorganisms for industrial applications with single-stranded DNA, enabling more efficient production of biofuels, pharmaceuticals, and other valuable chemicals. This innovative approach supports the development of sustainable and scalable biotechnologies.
Sources
1. Yoshimi, K., Kunihiro, Y., Kaneko, T., Nagahora, H., Voigt, B., & Mashimo, T. (2016). ssODN-mediated knock-in with CRISPR-Cas for large genomic regions in zygotes. Nature Communications, 7, 10431. https://doi.org/10.1038/ncomms10431
2. Li, H., Beckman, K. A., Pessino, V., et al. (2019). Design and specificity of long ssDNA donors for CRISPR-based knock-in. bioRxiv, 178905. https://doi.org/10.1101/178905
3. Bai, H., Liu, L., An, K., et al. (2020). CRISPR/Cas9-mediated precise genome modification by a long ssDNA template in zebrafish. BMC Genomics, 21,67. https://doi.org/10.1186/s12864-020-6493-4
4. Mason, D. M., Weber, C. R., Parola, C., et al. (2018). High-throughput antibody engineering in mammalian cells by CRISPR/Cas9-mediated homology-directed mutagenesis. Nucleic Acids Research, 46(14), 7436-7449. https://doi.org/10.1093/nar/gky550
5. Quadros, R. M., Miura, H., Harms, D. W., et al. (2017). Easi-CRISPR: a robust method for one-step generation of mice carrying conditional and insertion alleles using long ssDNA donors and CRISPR ribonucleoproteins. Genome Biology, 18, 92. https://doi.org/10.1186/s13059-017-1220-4
6. Miura, H., Quadros, R. M., Gurumurthy, C. B., et al. (2018). Easi-CRISPR for creating knock-in and conditional knockout mouse models using long ssDNA donors. Nature Protocols, 13(1), 195-215. https://doi.org/10.1038/nprot.2017.153
7. Roth, T. L., Puig-Saus, C., Yu, R., et al. (2018). Reprogramming human T cell function and specificity with non-viral genome targeting. Nature, 559, 405–409. https://doi.org/10.1038/s41586-018-0326-5.